library(palmerpenguins)
lm(bill_depth_mm~1, penguins)
Call:
lm(formula = bill_depth_mm ~ 1, data = penguins)
Coefficients:
(Intercept)
17.15 
Links to: Summary. Questions. Glossary. R functions. R packages. More resources.
Linear models provide a unified framework for estimating the expected value (i.e., the conditional mean) of a numeric response variable as a function of one or more explanatory variables. These models are additive: the expected value is found by summing components of the model — the intercept plus the effect of each variable multiplied by its value. The sum of squared differences between observed values and predicted values describes how closely the data match the model’s predictions. Linear models can be descriptive tools that capture the structure, variation, and relationships in a dataset. In later chapters, we will build on this foundation to evaluate models more critically — assessing how well they fit, how reliable their predictions are, and how to diagnose their limitations.
Try these questions! By using the R environment you can work without leaving this “book”. To help you jump right into thinking and analysis, I have loaded the ril data, cleaned it some, an have started some of the code!
R code and output below. The (Intercept) describes:
library(palmerpenguins)
lm(bill_depth_mm~1, penguins)
Call:
lm(formula = bill_depth_mm ~ 1, data = penguins)
Coefficients:
(Intercept)
17.15 Q3) Without running this code, predict which sex will be the reference level in the model: lm(bill_length_mm ~ sex, penguins)?
Q4 - Q7) Linear models with categorical predictors. The penguins data has data from three species of penguins – Adelie, Chinstrap and Gentoo. To answer the following questions, consider the R code and output below and no other information (that is, do not load these data into R).
lm(bill_length_mm~species, penguins)
Call:
lm(formula = bill_length_mm ~ species, data = penguins)
Coefficients:
(Intercept) speciesChinstrap speciesGentoo
38.791 10.042 8.713 Q4) What is the mean bill length of Adelie penguins in the dataset?
Q5) What is the mean bill length of Chinstrap penguins in the dataset?
Q6) How many mm longer Chinstrap bills as compared to Gentoo bills in this dataset?
Q7) What is the mean bill length of all penguins in this dataset?
Q8 - Q13) Mathematics of linear regression. Use the summaries below to conduct a linear regression that models the response variable, bill depth, as a function of the explanatory variable, bill length.
| mean_depth | mean_length | cov_length | sd_depth | sd_length |
|---|---|---|---|---|
| 17.59 | 46.57 | 0.62 | 0.78 | 3.11 |
| min_depth | max_depth | min_length | max_length | sample_size |
|---|---|---|---|---|
| 16.4 | 19.4 | 40.9 | 58 | 34 |
Q8) The correlation between these variables is .
Q9) The slope in this model is .
Q10) The intercept in this model is .
Q11) According to the model, what is the predicted bill depth (in mm) for a penguin with a 50 mm long bill .
Q12) A penguin with a 20 mm deep and 50 mm long bill will have a residual of mm.
Q13) According to the model, what is the predicted bill depth for a penguin with a 500 mm long bill mm deep bill.
Of course you haven’t!! The correct answer, of course, is that you cannot predict outside the range of our data. See Figure 1!
Q14 - Q16) More than one explanatory variable. Use the summaries below to conduct a linear regression that models the response variable, bill depth, as a function of the explanatory variable, bill length.
library(ggplot2)
library(dplyr)
library(palmerpenguins)
library(broom)
gentoo_data <- penguins |>
filter(species == "Gentoo")
lm(bill_depth_mm ~ bill_length_mm +sex, data = gentoo_data)|>
augment() |>
ggplot(aes(x=bill_length_mm , y=bill_depth_mm,color = sex))+
geom_point(size = 5, alpha = .7)+
geom_smooth(method = "lm",se = FALSE, linetype = "dashed",linewidth = 2)+
geom_smooth(aes(y = .fitted), se=FALSE, linewidth = 2)+
theme(legend.position = "top",
axis.title = element_text(size = 28),
axis.text = element_text(size = 28),
legend.text = element_text(size = 28),
legend.title = element_text(size = 28))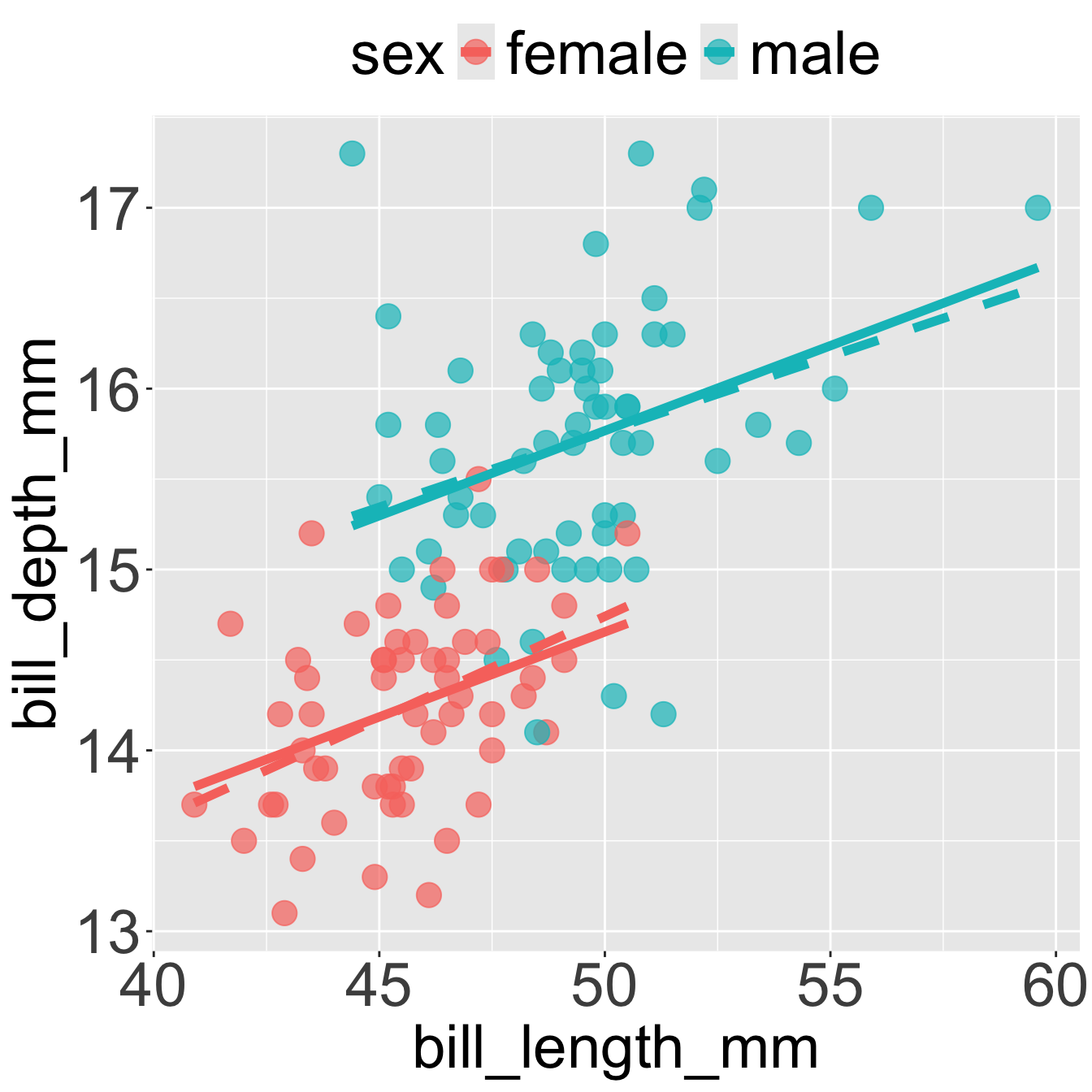
Q14) In Figure 2 there is a male penguin with a bill that is about 56 mm long (the second most extreme right point). Approximate, by eye its residual.
Q15) Based on Figure 2, which of the following are nearly identical for male and female Gentoo penguins? (Select all that apply.).
Q16) Use the web R space above to model flipper length as a function of body mass and sex of Chinstrap penguins. The sum of squared residuals is:.
Categorical Predictor: A variable with discrete groups. Modeled by differences in intercepts across groups.
Numeric Predictor: A continuous variable. Modeled by slopes showing expected change in the response per unit change in the predictor.
Indicator Variable: A numeric coding of a categorical variable (e.g., 0 for “pink,” 1 for “white”).
Reference Level: The baseline category in a categorical predictor against which other groups are compared.
b₀): The expected value of the response when all explanatory variables are zero (sometimes called \(a\)).b₁): The expected change \(\hat{y}_i\) with change in the predictor.
yᵢ): The actual value of the response variable: \(y_i = \hat{y}_i +e_i\).eᵢ): The difference between an observed value and its model-predicted value, \(e_i = y_i-\hat{y}_i\).lm(): Fits linear models. Syntax: lm(response ~ explanatory, data = dataset).augment() ([broom]): Adds predictions and residuals to your dataset for easy exploration.